One step closer to our goal: lab work
In my previous blog I wrote about playing undercover agent and purchasing all kinds of fish samples for DNA testing. This part was really fun, but I was also looking forward to the next phase of the project, which would get us closer to finding out the identity of our samples. For this phase it was time to get into the lab.

Some of the different seafood products we purchased to check whether they were correctly labelled. The samples include, according to their labels, grouper and bass. Photo © Ana Hacohen
The original plan was to send the samples to Mexico for DNA analysis. However, sometimes plans need to change and be adjusted to the circumstances. It turns out that transporting samples between countries can be quite complicated and frustrating. For example, in June last year I sent some shark samples from another project for DNA analysis. Although all the paperwork was in order, the samples were detained before they reached their final destination and we never got them back. So after a few Skype conversation with Adrian Munguia-Vega, my collaborator in Mexico, we came up with a new plan: we would do some of the laboratory analysis in Guatemala, specifically extracting DNA and amplifying it by means of polymerase chain reaction (PCR). Because we would not be transporting tissue samples, we expected the PCR products to arrive safely at the destinations where they would be sequenced.
However, we needed to find a laboratory with the necessary equipment to do this. After a few e-mails and phone conversations, the Biology Department of Universidad del Valle of Guatemala provided us with the space and equipment we needed. We were set! Adrian arrived in Guatemala a few weeks later and we were ready for two busy weeks in the laboratory when we would extract DNA from all our samples, solving any obstacles to the lab protocol on the fly.
The first part of the lab work involved cutting a super-tiny (1mm3) piece of tissue from each sample and placing it in a 2ml tube. It is very important to keep a good record of the samples, so we were careful to ensure that each tube was properly labelled. We then extracted DNA by using an enzyme that digested the tiny piece of tissue, releasing the DNA from its cells. To cut a long story short, the result was a new 2ml tube containing a few drops of clear liquid that we used for making millions of copies of the barcode gene we were interested in (COI) with the help of a PCR machine.

Placing samples in the PCR machine. This equipment is commonly used to amplify segments of DNA. Photo © Adrian Munguía
Next we needed to know whether the DNA amplification had been successful. To check DNA quality and quantity, we used an electric current to run DNA fragments on a special gel that can be seen under an ultraviolet light.

Adrian preparing the gel to check DNA quality. Photo © Ana Hacohen
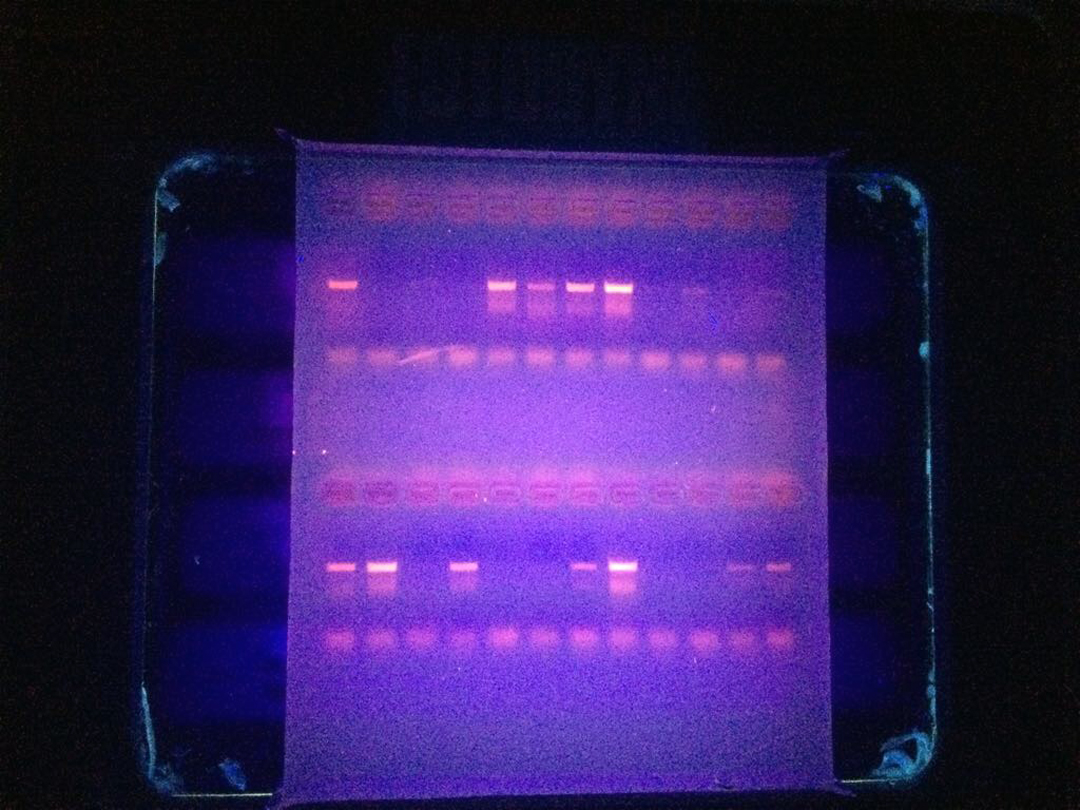
The gel seen under UV light. Photo © Adrian Munguía
Unfortunately some of our samples did not produce good DNA. We believe this may be because of the presence of different species in the samples, the presentation of the samples (fresh, frozen or dried and salted) and probably how the samples had been preserved during their journey along the market chain. However, we had collected some extra samples in anticipation of this happening.

The final product: a 96-well plate containing the DNA from our samples. Photo © Ana Hacohen
The last part of our lab work comprised waiting a few days for the samples to dry, resulting in a tiny amount of transparent film at the bottom of the tube. Finally, our PCR products will be sequenced in the laboratory. I can’t wait to find out which elasmobranchs are being sold and consumed in Guatemala and to see where the country stands in terms of seafood labelling.
