Sequencing Shark DNA

Photo © sirtravelalot | Shutterstock
To assess how distinct blacktip sharks from different countries in the Gulf of Mexico and Caribbean are we need to examine their DNA. In recent years, major technological advances have enabled geneticists to examine differences in DNA of many individuals across the complete collection of their genes, known as the genome. By examining differences in genomes of blacktips from different countries, we can find out if blacktips are moving across international boundaries and mating with sharks from other regions. This can tell us how best to manage blacktip populations and if international collaboration is required.
To examine differences in blacktip genomes, we need to sequence DNA. Preparing and sequencing a genomic library, a randomised collection of thousands of genes from hundreds of individuals, starts with shark fin tissue and ends with dozens of gigabytes of DNA sequence data. In this blog post, I detail a few of the main steps in this process. Let’s put on our white coats and enter the lab.

Dominic cutting up fin clips to extract genomic DNA. Photo © Dominic Swift
First, we need to extract DNA from shark tissue (Image 1). The best and least harmful way to collect tissue is a fin clip, a 1-inch piece of tissue taken from a shark fin. After collection, fin clips are placed in a buffer inside a small tube to preserve the DNA (Image 2). To extract DNA, we cut the fin clip into small pieces and place them in a buffer solution with an enzyme to break down the tissue and release DNA from the cells. After a few hours, specialised microscopic beads which bind to DNA are added to the solution, allowing us to isolate and collect DNA.

A blacktip shark fin clip stored in buffer to preserve DNA. Photo © Dominic Swift
Each genomic library contains DNA from up to 192 blacktips. To ensure we are examining differences in the same genes across all blacktips, we need to sequence the same sections of their genomes. To do this, we digest the genome of each shark into thousands of fragments using enzymes which locate sites of specific DNA sequence and cut the DNA backbone at these sites. The resulting fragments contain ‘sticky ends’, regions of DNA which can be glued to barcode sequences that allow us to identify which shark the fragments belong to. Barcoded DNA fragments from roughly 50 blacktips can be collected in a few drops of water and stored in a single tube, meaning a complete genomic library of 192 blacktips can be prepared in four tubes (Image 3). These tubes are then mailed to a facility which sequences the DNA code for each fragment in all blacktips. A few weeks later, approximately 60 gigabytes of DNA sequencing data are downloaded, allowing us to start examining genomic differences in blacktips from the Gulf and Caribbean.
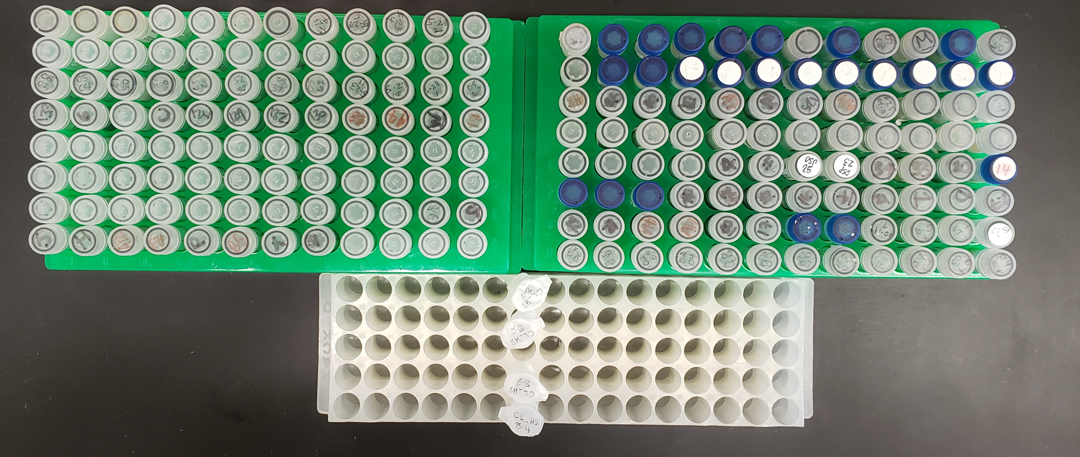
Tubes containing fin clips from 192 blacktips and 4 tubes containing DNA from each of these blacktips. Photo © Dominic Swift
