Unwinding the Helix: Extracting Genetic Relationships from Nurse Shark DNA in The Bahamas
There are many unique features that make you who you are. If you were to describe them to somebody, you would probably include your personality, some physical characteristics, and the people that you surround yourself with. However prominent, there is a piece that you will probably forget to mention: your DNA.
DNA (deoxyribonucleic acid) holds the code to every facet of our being, from what makes our hair curly or straight, to what colour our eyes are, to the pigment of our skin. Our bodies read our DNA as a recipe for creating the wonderful makeup of you. No matter if we view our DNA as a curse or blessing, one thing is undeniable: it is something we cannot escape from.
It can often be challenging to pick out unique individuals within the same species. To us, some of the individual features may not be as distinguishing as it can seem in humans. A surefire way to differentiate individuals is by using their DNA. Depending on the type of organism, scientists can take a tissue sample that will hold the individual’s DNA or collect samples that the individual has left behind in their environment.
What does this look like in sharks? Scientists perform a “shark workup” that is akin to a shark version of a doctor’s visit. As part of this, a small piece of tissue (typically from a fin) is collected and stored in a vial with a preserving agent such as ethanol or DMSO. This fin clip contains dermal denticles and cartilage tissues that hold the individual shark’s DNA, which contains a suite of information used to address various research questions.

While doing a shark workup on a juvenile lemon shark (Negaprion brevirostris), the BBFSF team removes a tiny piece of fin to store for DNA analysis. Photo © Chelle Blais
Monitoring sharks is a lot more difficult compared to their terrestrial counterparts, so genetic analysis can be a powerful tool to provide insight into their life history and relatedness. Similar to humans, if an individual shark shares a significant portion of its DNA with other individuals, there is a high probability that these individuals are related. If they were captured near the same location, it is possible to further deduce that these individuals are not moving far and/or a parent has site attachment to where they give birth. Conversely, if closely related individuals were captured farther apart, this is a good indication that these sharks are traveling long distances. This information can help inform population structure and connectivity within a species, which has important implications for management and conservation. Using shark DNA has also been informative for determining hybridisation (offspring from the result of two different species successfully mating and reproducing), parthenogenesis (females giving birth without a male mate), long-term sperm storage in females, gene adaptations to changing environmental conditions, and natal philopatry (the return of females to their birthplace to give birth to their pups).
Nurse sharks (Ginglymostoma cirratum) are an abundant, coastal, shallow water species found throughout the Atlantic Ocean and were listed as ‘vulnerable’ by the IUCN in 2021. There is limited major research on nurse sharks in The Bahamas, but The Bimini Biological Field Station Foundation (BBFSF – Shark Lab) located in Bimini, The Bahamas, has been collecting data on the species since 1990. As part of this research, the Shark Lab has historically collected fin clip samples, which now provide an underutilised sample bank for DNA analysis (Figure 2). Fast forward through 30 years of sample collection by the Shark Lab and 350 nurse shark tissues later, and this is where I come in! For my master’s project at the University of Nebraska-Lincoln (UNL), I am using nurse shark DNA to help improve our knowledge on the local and regional population structure and connectivity of this species around Bimini and throughout The Bahamas.
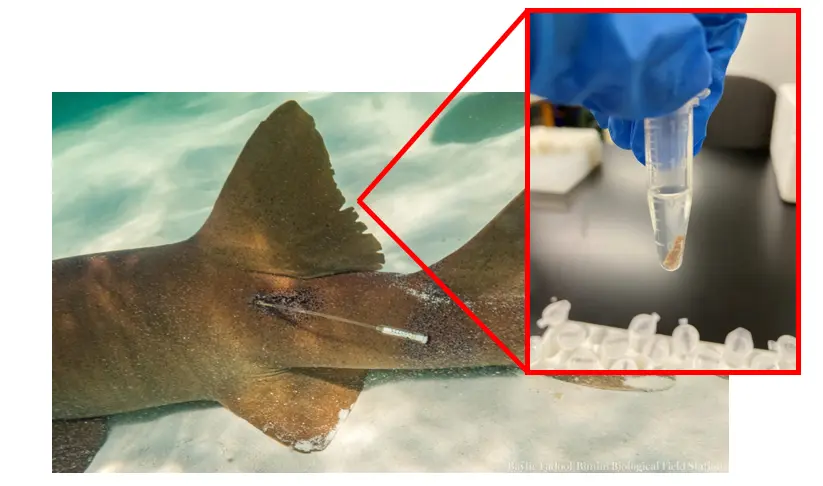
A fin clip from a nurse shark (Ginglymostoma cirratum) in Bimini that will be used for assessing the population structure and connectivity of nurse sharks in The Bahamas. Photo © Baylie Fadool
After the completion of field work, I transferred all the nurse shark samples to the Field Museum in Chicago to begin the DNA extraction process (which has ended up being one of my favourite parts). For this first step, it is as simple as taking the nurse shark fin clip and cleaning it from all other materials to leave only the DNA from the individual behind. Below, you can see me in the middle of this process which took about three weeks for 350 samples.

Baylie beginning the DNA extraction process at the Field Museum in Chicago by subsampling nurse shark (Ginglymostoma cirratum) fin clips. Photo © Toni Muzzo
Once the first step was completed, I brought all my DNA extractions back with me to UNL. This next portion can be completed following a multitude of different laboratory methods depending on the question that you want to answer. I am interested in cryptic, fine-scale population structure, and to do this, I am following a protocol called double digest restriction site-associated DNA sequencing (ddRAD-seq) and using a genetic marker type called single nucleotide polymorphisms, or SNPs, that are highly variable throughout an individual’s genome (the complete set of all the shark’s DNA).
To visualise what laboratory methods need to be done next, imagine walking through a library looking for a specific book. What information would you need to find this book? Perhaps the title of the book would be the most important piece of information. Let’s call the title of the book the unique index. Each nurse shark sample is prepared the same way by receiving a unique index that helps me identify which individual is which. No two indices are the same just as no two title and author combinations of a book are the same. Once my individuals receive their index, I select a size fragment for the DNA, I purify it, and then I pool them all together into one big DNA soup (the most unique of alphabet soups). This is where the indices I gave each individual are important, as I use these to distinguish between individuals in this DNA soup after sequencing. Sequencing is like the reader in the library, while the DNA is the book. The sequencer reads the DNA and then writes a summary on the DNA. Imagine me as the librarian, as I then take on the task of interpreting the DNA summary.
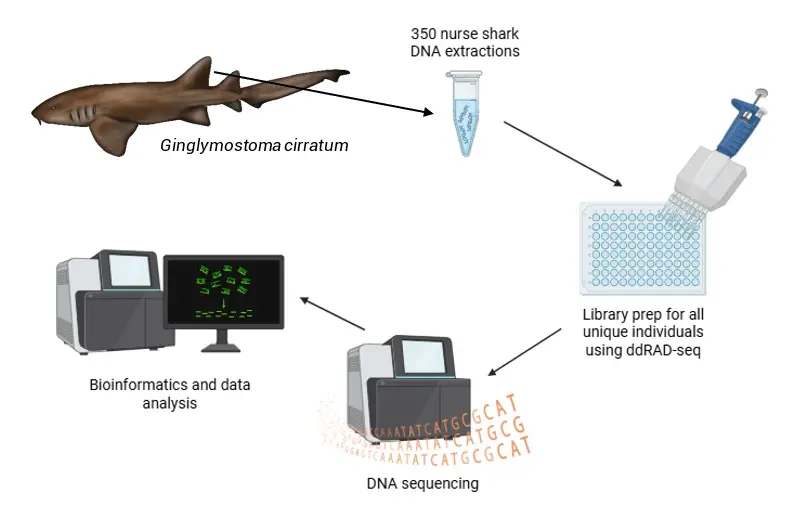
Diagram of the laboratory methods for DNA sequencing in nurse sharks (Ginglymostoma cirratum). Nurse shark illustration by Chelle Blais. Figure © Baylie Fadool using BioRender
Working in genetics can sometimes sound scary and make you immediately imagine scientists in white lab coats conducting precise experiments with hair standing straight up to the sky. Although my hair usually does fly in all directions, mostly due to its inability to cooperate with me on any given day (but hey, that’s just my genetics), the information can really be broken down to make sense to anyone! I love that the laboratory work can be done from anywhere, as you do not have to be located close to the ocean to be working with shark DNA. The repetition of genetics work also provides a platform to hone a wide range of laboratory skills and confidence in analytical skills, in addition to the field work experiences I have had. I am extremely excited for the next stage of my master’s project, which is interpreting the genetic data and putting this in the broader context for better understanding the life history of nurse sharks. Stay tuned for many presentations and publications in the future that will help contribute to the knowledge we have of this abundant but still ‘vulnerable’ species.
