Preparing to reliably find needles in the oceanic haystack
A lot has happened since our last blog post! A big reason for the gap is because it can take a year or more to develop appropriate procedures and protocols for conducting reliable environmental DNA (eDNA) research. Contrary to what some people think, simply taking a water sample, sending it to a genetics lab, and finding out if a species was detected is NOT how it works. As in many aspects of life, if something seems too easy to be true, it probably is.
For example, since the beginning of the project, we’ve moved from using quantitative polymerase chain reaction (qPCR) techniques to droplet digital (ddPCR) techniques. For most people, this change likely doesn’t mean much, but when you’re looking for tiny fragments of DNA in a huge ocean, it’s a big deal!

We’re looking for tiny fragments of DNA in a huge ocean, so essentially we’re looking for the proverbial needle in a haystack! Nicole Phillips (seated) and Rachel Scharer filtering samples. Photo © Gregg Poulakis.
This analytical shift was made possible by communication with our collaborators in Dr. Nicole Phillips’ laboratory at the University of Southern Mississippi. It turns out that Nicole and her graduate student Ryan Lehman were asking similar questions about the smalltooth sawfish in waters off Mississippi as we were in Florida. Together, we have been working on techniques to avoid “false negatives” and “false positives” before embarking on any field work in areas outside the current range where this Critically Endangered species might be found.
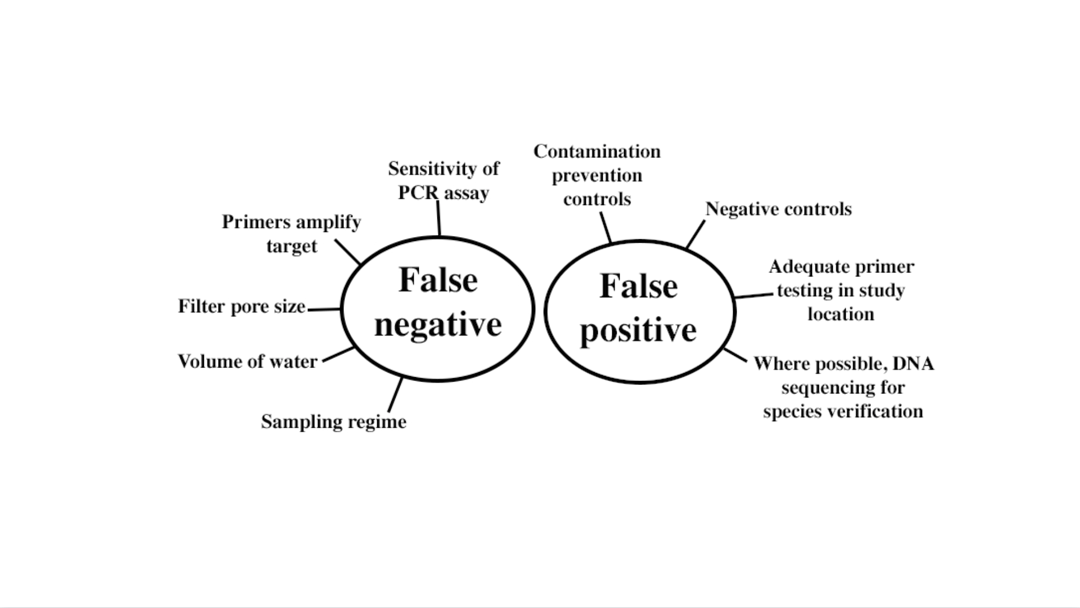
Two things that any eDNA study wants to avoid are false negatives and false positives. Both things are influenced by multiple factors. Figure provided by Nicole Phillips.
One of the biggest concerns in a study like this is what’s called a “false negative”. This means that DNA from your target organism was in your water sample, but your PCR assay wasn’t sensitive enough to detect it (see other considerations in the figure). To avoid this, you want to use the most sensitive assay possible, especially if your species is rare, and currently, the most sensitive assay is ddPCR. The smalltooth sawfish assay our team is using can detect < 1 picogram of target DNA in a sample; 1 picogram is 1 trillionth of a gram! Assays for different species will have different detection capabilities depending on the primers and how well optimized they are.
Since Nicole’s lab can use ddPCR to analyze the samples, next we needed to address another potential issue, “false positives”. To do this, we developed and thoroughly tested a primer set and probe, so we are 100% sure that any “positives” indicate the true presence of smalltooth sawfish and not any other closely-related species that share the same waters such as guitarfish or stingrays. That was said in one sentence, but the process took months of work! Luckily, archived fin clip samples from many years of sampling helped speed up the optimization process somewhat. The good (great!) news is that we have a primer-probe set that works in our study areas, so our field positives will definitely represent the smalltooth sawfish!
Over the last year, in addition to primer-probe set development, we’ve settled on our experimental design that includes some randomly generated sample locations, some non-random sample locations based on sawfish encounter reports reported by the public, bleaching everything (to avoid false positives from contamination), a nylon filter pore size of 0.8 micrometers (small, but we want to minimize missing DNA), a bottom-water volume of 3 liters (sawfish live on the bottom and we want to filter enough water to minimize false negatives), and you guessed it, bleaching everything yet again (our slogan is “when in doubt, bleach it out!”).
That’s it for now, but look for updates in upcoming blogs on: Defining what positives mean and the importance of collaboration and communication.
