Preparing to analyze sawfish environmental DNA
We enter the lab and can smell nothing but progress—defined as the lingering odor of fecal samples—thawed that morning to conduct an experiment. These samples were opportunistically collected during field research on the Critically Endangered smalltooth sawfish in southwest Florida, USA. Unfortunately, while what we called progress was waiting to be processed, researchers using this shared lab space were not quite as enthusiastic about the fragrance we had invited upon the room. Becoming social pariahs among our colleagues is just one of the many perils faced when conducting this research. It may sound odd, but these rare fecal samples hold the keys to important research and conservation questions because about 90% of these samples are composed of smalltooth sawfish DNA!

Smalltooth sawfish feces opportunistically obtained and stored in 20 ml scintillation vials by the Florida Fish and Wildlife Conservation Commission during ongoing monitoring and field research. Photo © Taylor Hancock
The experiment we conducted was an effort to examine the properties of one hypothesized source of environmental DNA (eDNA), which is loose or particle-bound DNA derived from shed biological materials such as skin cells or the products of reproduction, decomposition, or defecation. eDNA is an emerging conservation technology which allows for the detection of an organism by analyzing DNA extracted from water or sediment samples. This detection method is appealing as the target organism does not have to be present when the samples are collected. This allows for a larger window of detection than other methods, and is helpful when attempting to detect rare or cryptic species, such as the smalltooth sawfish. Our aim for this experiment was to guide us in the development of a smalltooth sawfish eDNA detection method in Florida.

Smalltooth sawfish feces suspended in sterilized ultrapure water to simulate eDNA. Photo © Taylor Hancock
Our experiment involved suspension of a portion of a fecal sample in sterilized water and then filtering it through successively smaller pore sizes. From here, we extracted the DNA caught by these filters and measured the concentration of all eDNA extracted, which showed that there was more eDNA present at the 5–10 µm particle size as compared to the 0.7 µm and 20–25 µm particle sizes. This was a promising result; however, this does not necessarily mean this reflects the abundance of the eDNA of our target organism. To measure this, we used a highly sensitive DNA quantification technique which allowed us to accurately measure the concentration of DNA of the smalltooth sawfish within the total extracted eDNA. This technique involves a quantitative polymerase chain reaction (qPCR) with the use of a primer set we developed with our collaborators, which amplifies a specified sequence of smalltooth sawfish DNA, and only smalltooth sawfish DNA. We determined that over 90% of the feces were trapped by the 20–25 µm and 5–10 µm filter fractions. We will use these results to continue developing our field protocols for this summer’s sampling in areas just outside the current range of the species.
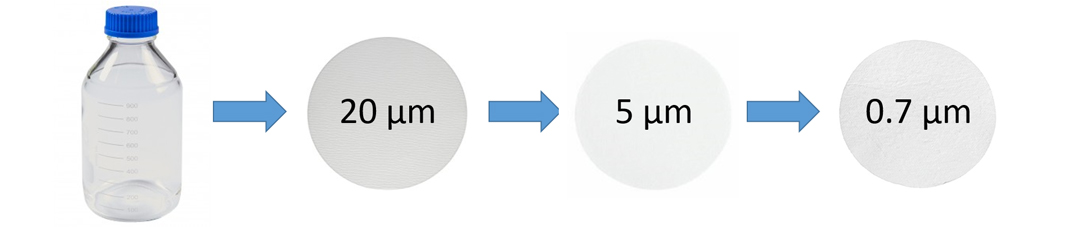
Generalized visual representation of the smalltooth sawfish fecal eDNA quantification experiment. Photo © Taylor Hancock
