Genetics to the rescue!
Assessing population connectivity in the critically endangered common eagle ray using DNA
– paraphrasing John Shepherd
Indeed, for us to know how many fishes are in the sea, we need to count them. It sounds like an easy task at first, it is just counting, right? But marine fishes have this uncanny ability to not be where we think they are, or to not bite when we need them to. There is also the added challenge that we cannot possibly sample every single fish in the sea. So how can we count, or more broadly speaking, how can we study something we cannot see?
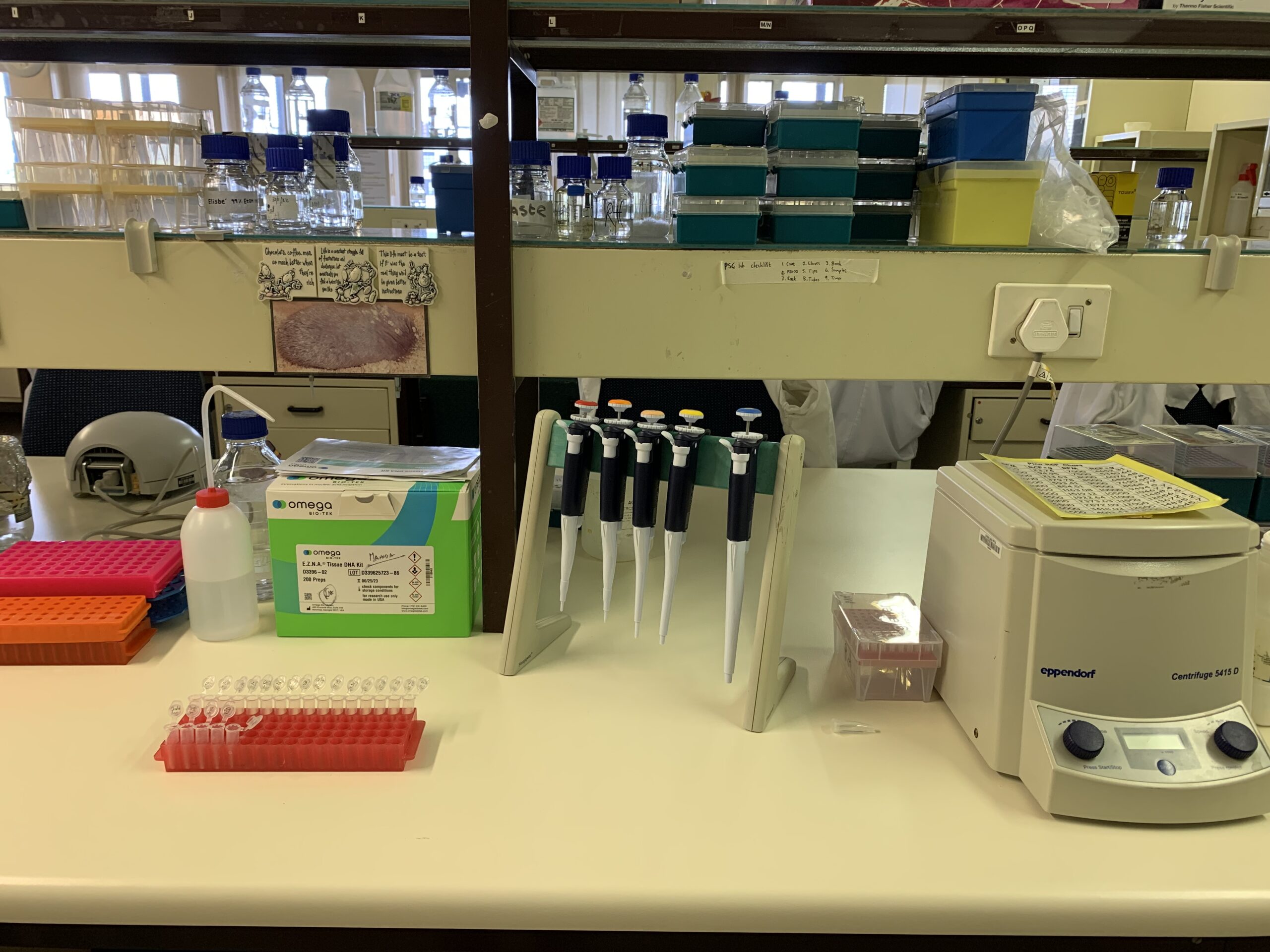
Marine genomics lab at the University of Pretoria. Photo © Romina Henriques
This is where genetics and molecular ecology can help us. By studying the DNA of fishes we can understand how many of them there are, how their populations are linked, or even how healthy these populations may be. Just like humans, each fish has its own DNA profile, which we can compare to others, to understand if they are all part of the same population or if there are many separate populations distributed throughout the world. And to do this, we just need a little bit of skin or muscle tissue, and a small sample of individuals – no need to fish all the fishes out!

Romina Henriques extracting DNA from tissue samples. Photo © Romina Henriques
One of the most interesting discoveries from molecular data was to debunk the long-held idea that fish could swim everywhere across the Oceans: their DNA is showing us that this is not always the case. Even though there are no obvious physical barriers to migration in the sea, and some fishes (especially sharks and rays – hello, shortfin mako sharks!) can swim very long distances, molecular evidence is mounting showing that many species are composed by multiple populations that haven’t been in contact with each other for many generations.
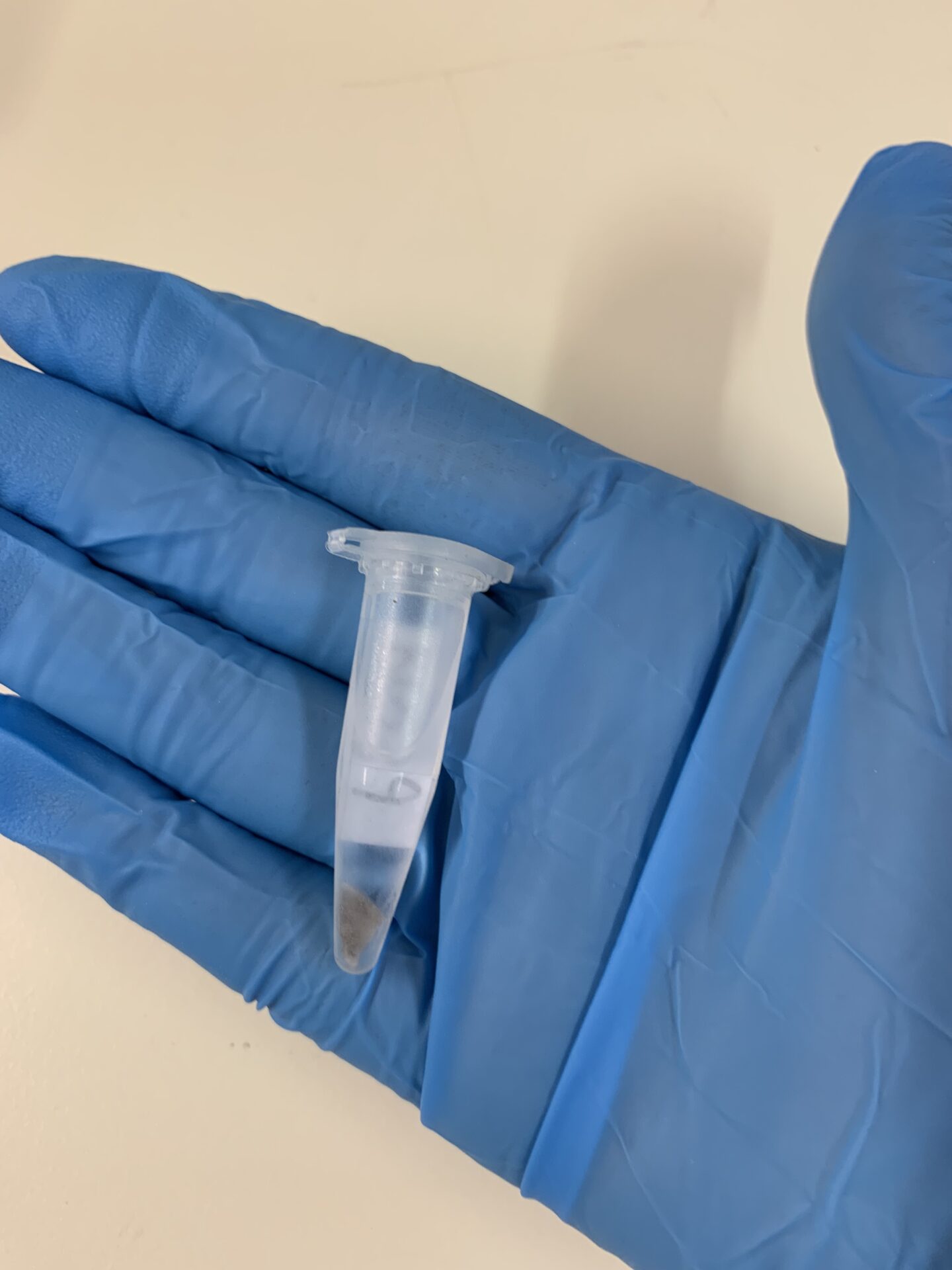
Example of a tissue sample for DNA extraction. Photo © Romina Henriques
In our SOSF-funded project, Unearthing the truth about the common eagle ray (Myliobatis aquila), one of our aims is to assess genetic connectivity across the southeastern Atlantic for this iconic and highly mobile species. We are going to use a combination of molecular tools, such as sequencing a gene of the mitochondrial DNA (which is only inherited via the mothers) as well as sequencing a portion of the entire genome, to understand how, and if, the common eagle rays from Angola and South Africa, as well as within South Africa, are connected. The common eagle ray is Critically Endangered, and thus it is fundamental to know how its populations are connected, to better establish effective conservation actions to protect it.
